AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling

AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling
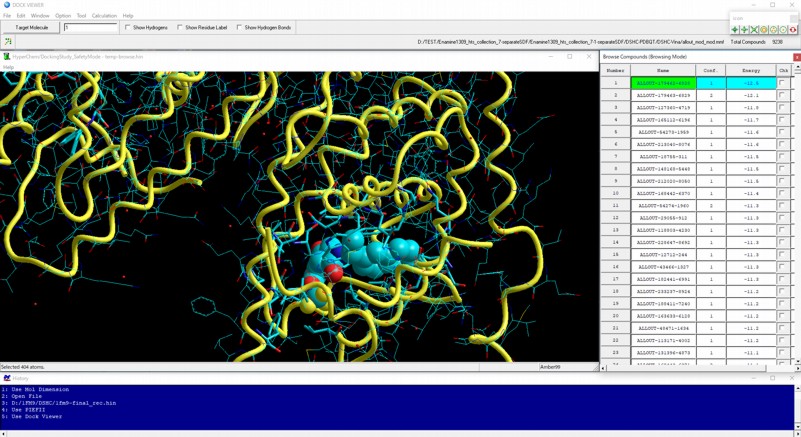
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -

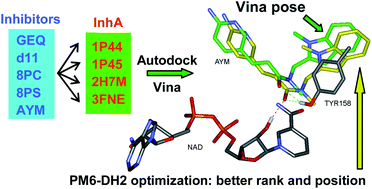
Optimization of Autodock vina program by docking the co-crystal ligand... | Download Scientific Diagram

How to install Gromacs, PyMOL, AutoDock Vina, VMD, MGLTools, Avogadro2, Open Babel in Ubuntu 20.04 - DEV Community

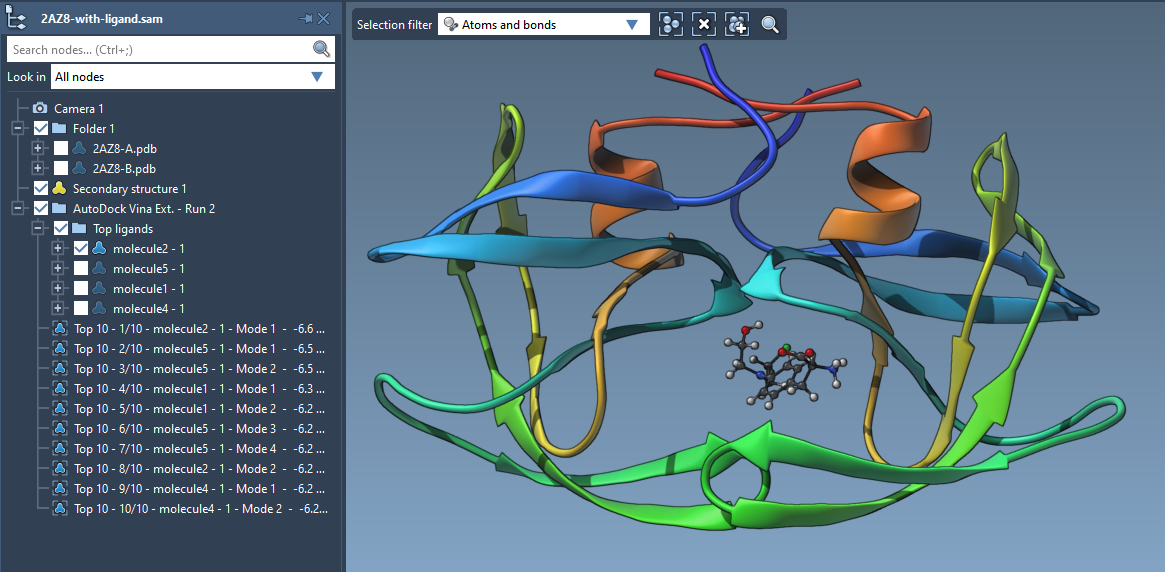
The new version of AutoDock Vina Extended is out for SAMSON 2020 R2 | The new version of AutoDock Vina Extended includes interactive analysis of docking results by linking plots to conformations.
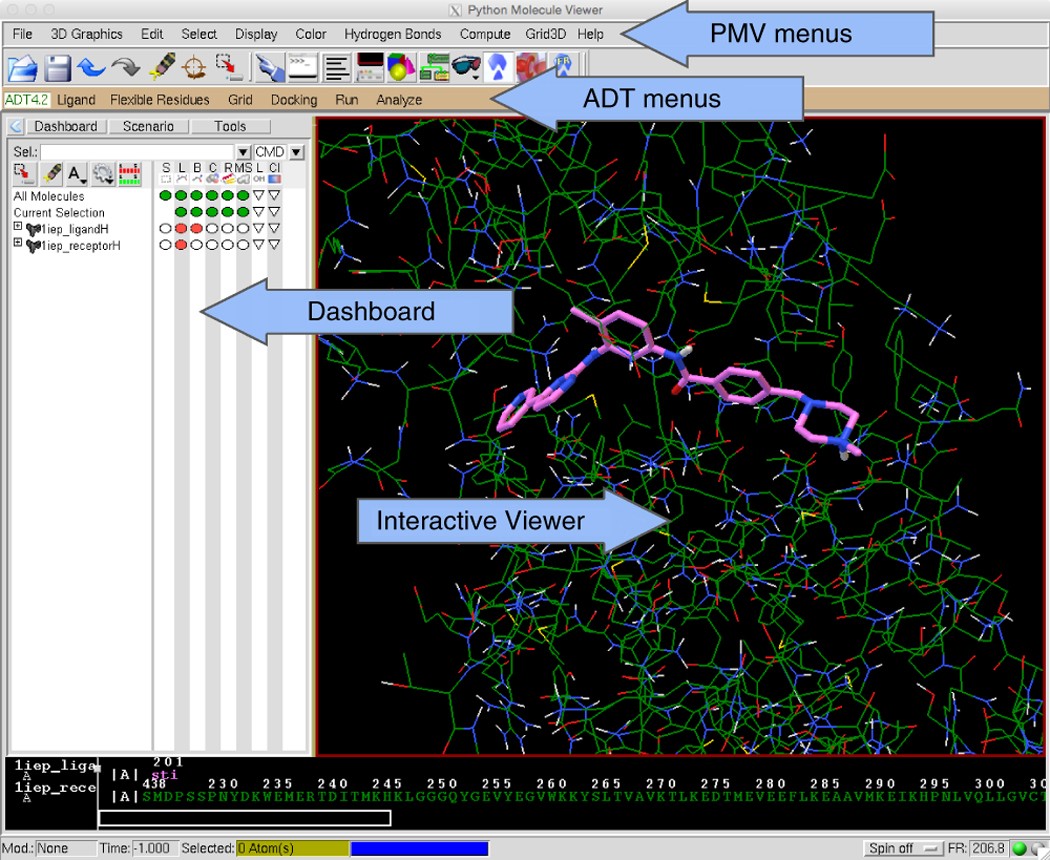
Computational protein–ligand docking and virtual drug screening with the AutoDock suite | Nature Protocols

SAMSON on Twitter: "AutoDock Vina in SAMSON: flexible protein-ligand docking in a few clicks http://t.co/lCyVMXt1nh" / Twitter

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect
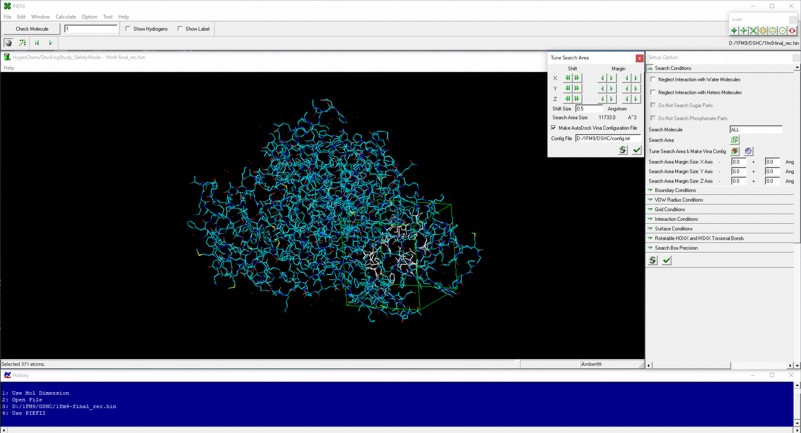
AutoDock Vina In Silico Screenings Interface: Docking Study with HyperChem - Institute of Molecular Function -

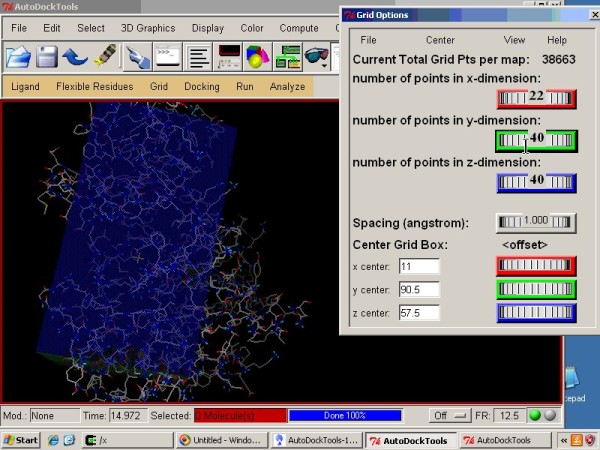
3 Autodock GUI with AutoDock-Vina by ADT. (a) Schematic representation... | Download Scientific Diagram